

- Hidden markov model matlab code for biome data how to#
- Hidden markov model matlab code for biome data trial#
- Hidden markov model matlab code for biome data series#
Hidden markov model matlab code for biome data how to#
These HMMs were thenused to both generate a generic template of activity, and also to recognizewhich pin was stimulated on a previously unseen data set.Īll HMMs wereimplemented using Kevin Murphy’s MATLAB toolbox, and many of our choicesregarding how to adapt HMMs to this application closely follow the work ofSeidemann et. An HMM is a statetransition model defined primarily by the number of states represented, thetransition probabilities between these states ( P ), and the likelihood of observing a particular signal given a certainstate ( O ). This spike data wasthen used to train an HMM for each stimulated pin, which succeeded inextracting the general dynamical properties of the trials. This data wasfirst filtered and spikes were extracted for each channel.

Hidden markov model matlab code for biome data trial#
Each of the 64 channels were sampled at a rate of 20 kHz.įigure 1: MEA pin layout highlighted pins are those whichwere stimulatedįigure 2: Single trial raw datafrom each of the 64 pins only the first 100 ms is shown Seven or eight trials were recorded for eachstimulation pin an example of the raw data for a single trial can be seen in Figure 2.
Hidden markov model matlab code for biome data series#
Cells werecultured on the array for several days, and then a series of experiments wereperformed in which pins in the second and seventh columns were stimulatedindividually (Figure 1). These arrays contain 64 electrodes in an 8-by-8square, with electrodes spaced 150 microns from each other.
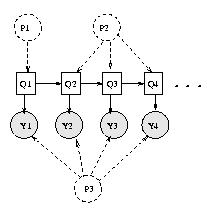
The data usedin this study was collected by Nathan Wilson using Med64 electrode arrays( ). What is the pattern of connectivity between thecultured neurons, and how does this connectivity relate to the observedsignals? Do the cells project axons across the entire extent of the array, ordo they connect preferentially to those cells which are nearby? Do thecultured networks produce stereotyped patterns of activity, or is this activitydiffuse and undifferentiated? The present study will attempt to answer thislast question. The largeamounts of data resulting from these recordings prompt many questions and posea great analytical challenge. Theelectrodes can then be used to record the spiking activity of the neurons, aswell as to stimulate those cells at certain locations on the array. Using the proper cell culturingtechniques, neurons can be induced to grow on the surface of the arrays. Multielectrode arrays (MEAs) aretwo-dimensional arrays of electrodes. Thissuggests that 1) stimulation of a particular pin invokes a stereotyped spatialand temporal pattern of activity, and 2) these patterns are distinct from one electrodeto the next. In addition, we show that appropriately trained HMMscan recognize which pin was stimulated given a previously unseen trial. We find that, given appropriate initialconditions, HMMs can be trained to recognize sharp borders between a number ofoverall firing states. We apply the Hidden Markov Model (HMM) framework in order to characterize theresulting activity patterns. The specific dataexamined here is the result of stimulating a single pin in the array at a time. While it is assumed that these cells formnetworks during their growth, little is known about the nature of thesenetworks or the dynamical properties of their activity. Multielectrode arrays allow the simultaneous recording ofthe activity of many cultured cells. Modeling theDynamics of Multielectrode Array Recordings Using Hidden Markov Models


 0 kommentar(er)
0 kommentar(er)
